One of the main objectives of the IDEA4RC project is to develop an algorithm capable of extracting data from clinicians’ notes and pathology or radiology reports stored within the hospital’s information system. Currently, a wealth of information is locked within these texts, which cannot be fully utilized by researchers to uncover more about rare cancers, how they can be diagnosed and treated, the factors influencing prognosis, and the effectiveness of treatments.
The data extracted from the texts can then be stored inside the IDEA4RC secure processing environment of each hospital and made available through the IDEA4RC ecosystem for researchers to conduct analyses on rare cancers.
Since the texts are written in natural language, IDEA4RC researchers can leverage recent advancements in natural language processing (NLP) algorithms, largely driven by deep learning techniques.
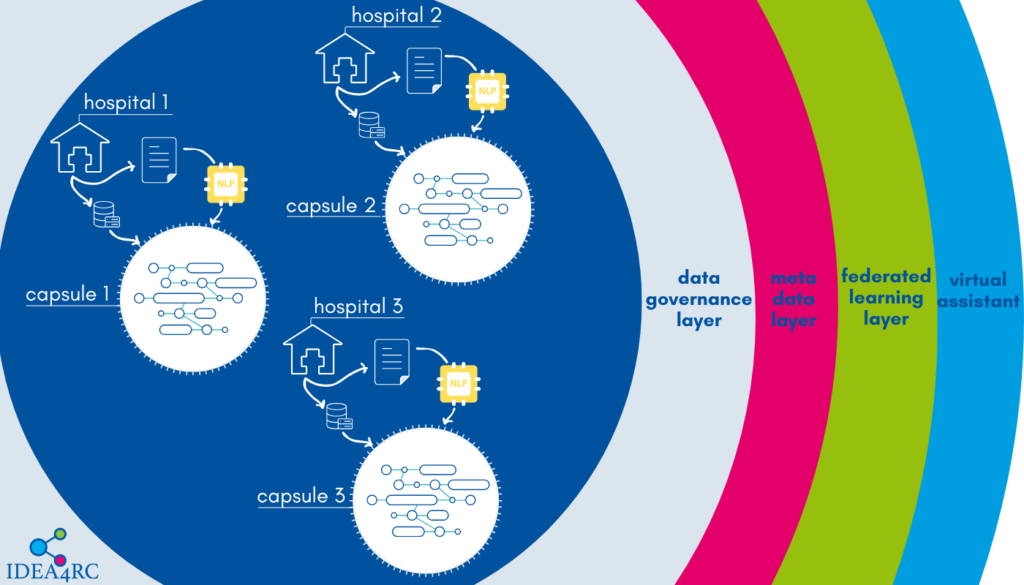
However, the task at hand is a challenging one. Extracting information from clinical notes and reports not only requires a deep understanding of language — a strength of Large Language Models (LLMs) like those powering ChatGPT or Gemini — but also demands specialized clinical knowledge.
Furthermore, while companies like OpenAI or Google can access vast datasets to train their models, high-quality clinical notes and pathology or radiology reports are limited, especially on rare cancers, and protected by privacy regulations. The model developed for the IDEA4RC project must also be multilingual, as the notes are written in the various languages spoken at the consortium’s participating centres.
We interviewed Unai Zulaika Zurimendi, a computer engineer at the University of Deusto in Bilbao, and Soumitra Ghosh, a researcher at Fondazione Bruno Kessler in Trento, who are leading the NLP activities within the IDEA4RC project.
Unai, why is it so important to be able to extract information from clinicians’ notes and pathology or radiology reports?
Our starting point were the IDEA4RC data models for head and neck cancers and soft tissue sarcomas that we developed during the first two years of the project by gathering the knowledge of clinicians and researchers working on rare cancers within the 11 clinical centres involved in the project.
The data model is essentially a list of variables logically related to one another that clinicians deem relevant to understand the disease. While some of these variables can be extracted from the databases maintained by the centres, others are only documented in the notes written by clinicians, surgeons, pathologists, and radiologists who follow the patients through the disease and associated treatments.
The first step was understanding which centres need to extract which information from texts. In some centres, there’s a significant amount of information locked inside these notes that we would not be able to exploit without a custom NLP model.
What are the next steps to deliver an algorithm capable of extracting information from clinical texts?
NLP model are machine learning models, which means they need to be trained on data. Given the highly specialised knowledge required to understand which part of a text corresponds to which variable in the rare cancer data model, we need to use supervised learning. This means that we need to start from one of the available Large Language Models and show to this algorithm many examples of association between expressions (words or group of words) in our clinical notes and values of specific variables.
How will you do this?
Clinicians at the participating centres will annotate the texts for a small subset of patients treated there. Annotating means they will highlight spans of text and indicate which variable can be assigned a value from that and which is this value.
To do this, they will use a tool developed by CliniNote, a Polish health data company and a partner in the project. This software is based on a tool that CliniNote has already commercialised, which has been adapted to the scope of our project. It allows clinicians to visualize all notes related to a specific patient in a single window and suggests which variables are required (those that cannot be derived from other already structured data sources at their hospital). Clinicians can then browse through the notes — whether they are visit records, pathology, or radiology reports, highlight relevant phrases, and use the software to generate a standardized sentence that only needs to be filled with specific values.
Using templated sentences is intended to streamline the annotation process for clinicians, as they are often already familiar with this format.
Can you give us an example?
Yes. For instance, suppose you wish to determine the stage of a soft tissue sarcoma from the clinician’s note below.

Following the TNM staging system for trunk and extremities soft tissues sarcomas, one would deduce that T is equal to two, since the tumour’s size is between five and ten centimetres, and that N and M are both zero, because there isn’t any lymph nodes involved, and the cancer did not spread to other parts of the body.
, and that N and M are both zero, because there isn’t any lymph nodes involved, and the cancer did not spread to other parts of the body.Another example would involve the determination of the sarcoma’s grade. The grade is a characteristic of the cancerous cells and can only be determined through a histopathological examination of the tumour’s tissue. The pathology report would look something like the text below.
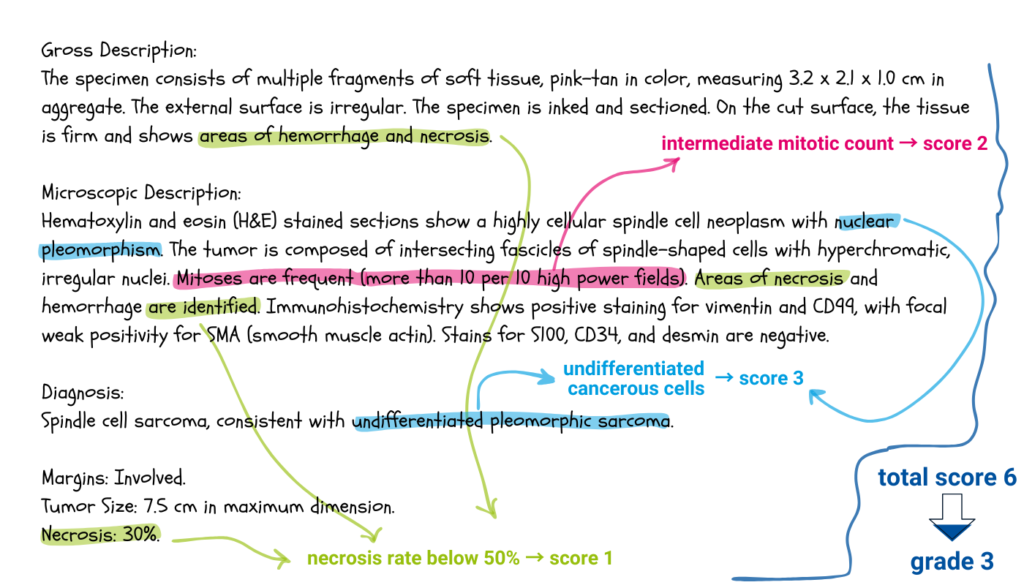
Following the ESMO guidelines, clinician can identify the scores associated to differentiation, mitotic count and necrosis rate that all together determine the grade of the tumour.
What kind of challenges has the annotation work entailed so far?
The challenges have been twofold. On one side, we needed to set up the annotation software to make it as user-friendly as possible for clinicians. Their time is valuable, so we must do our best to ensure that annotation is as efficient and accurate as possible.
On the other side, there were legal issues that we needed to resolve. We established and signed agreements between each clinical centre and Fondazione Bruno Kessler to allow the transfer of annotated texts to Fondazione Bruno Kessler’s servers, where the actual training of the model will take place. The training phase requires high computational power, particularly the availability of graphics processing units (GPUs), which most clinical centres do not have.
Soumitra, what are the challenges of training such a model?
We are navigating uncharted waters. The task we need to accomplish is quite specific and our dataset is a real-world one. So far, researchers have been developing models in a very controlled environment, with well curated datasets. Most of these models perform a task called Named Entity Recognition.
When performing Named Entity Recognition, a model is presented with a sentence like the following:
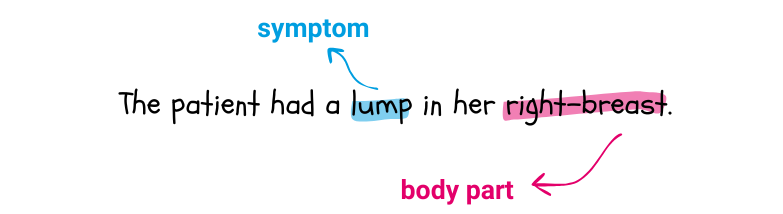
and is asked to identify the terms in the sentence that refer to some predefined clinical entities. In this case, it could identify “lump” as being a symptom and “right-breast” a body part. Symptom and body part are two clinical entities.
Instead, we need to address the specificity of the cancer models developed by IDEA4RC and account the great variability in the style and format of our dataset.
Moreover, we are interested in associating a date with each event or variable’s value, as researchers are interested in the temporal dependencies among variables. It may well happen that to understand the stage a tumour at diagnosis, clinicians must refer to more than one note and combine the information.
An additional challenge will be setting benchmarks, which means establishing when we can consider ourselves satisfied with the model’s performance. There is currently no literature on this type of task.
How will you train the model?
We will begin with the multilingual large language model developed by Meta, called LLaMA. LLaMA is a deep neural network pre-trained on a vast number of texts, sourced from the internet and digitized libraries, to understand the structure of natural language. Meta has made this model available to researchers. From there, we will fine-tune the model using the annotated texts we will receive from the participating centres.
After training the model, what will be the next steps?
We will test our model on a fraction of the annotated texts that we won’t use for training. If the test results are promising, we will deploy the model on the remaining patient notes selected by the centres, extracting data and injecting it into the capsules that populate the IDEA4RC data ecosystem.
Natural Language Processing will also be employed for a second purpose in the project. Can you tell us what that is?
Yes, NLP will also be used to develop a question-answering system to assist IDEA4RC users in exploring the data and building patient cohorts more effectively.
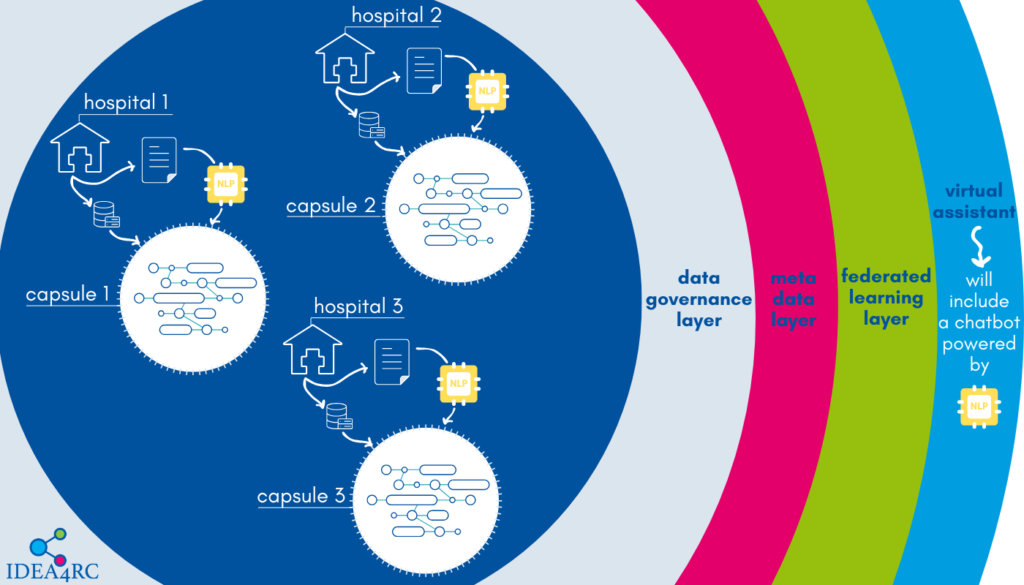
For instance, if a researcher wants to build a cohort of patients diagnosed with a specific type of cancer during a given period, they can type their query in natural language. The algorithm will then generate the corresponding SQL code to retrieve the desired cohort, as shown in the example below. This feature will simplify interactions with the IDEA4RC data ecosystem significantly.

